| Visitors Now: | |
| Total Visits: | |
| Total Stories: |

| Story Views | |
| Now: | |
| Last Hour: | |
| Last 24 Hours: | |
| Total: | |
Nanopore sequencing: The next big thing?
The biotech field is abuzz with nanopore sequencing, with lots of people starting to think it might be the Next Big Thing. And not just one of those incremental developments that keep science humming along, but more along the lines of Kuhn’s paradigm shifts. The belief is that nanopore sequencing may finally lead to the domestication of biotechnology, which will open up brave new worlds in all kinds of domains like drug development, law and health insurance. The ultimate manifestation of this technology, of which Oxford Nanopore Technologies is the leader, is a flash drive that can be plugged into your computer and which can sequence your DNA for about 1000$ within a day or two. Personal genomics does not get any more well-defined than that.
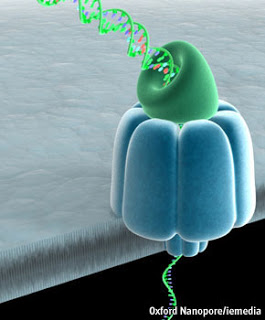
There’s an article on nanopores in this week’s Science which lays out both the promises and the challenges. Nanopore sequencing seems to be one of those disarmingly simple ideas which had to be hammered out into practice by sweating all the details. About half a dozen people, starting with David Deamer at UCSC, contributed to the basic concept which sounds simple enough; find a pore with an electrical current flowing through it and thread a DNA strand through the pore. The expectation is that as each base of the DNA passes through, there will be a small, characteristic change in the ionic current. The great hope is that this change would be unique for each one of the four bases, thus allowing you to read the sequence.
As usual, the implementation of the idea faced major hurdles. For one thing, nobody knew of a pore that would allow DNA to thread through it until Deamer heard of alpha-hemolysin, a protein with a magnificent structure that is used by some bacteria to break down red blood cells by literally drilling holes in them. Deamer realized that the perfectly positioned inner channel of hemolysin could be used to string DNA through it. After this initial breakthrough, the main problem remained reducing both the number and the speed of the base pairs enough to read them accurately. The first problem was resolved through the discovery of another protein called MspA which has a pore narrow enough to allow just four base pairs through at any given moment. The negatively charged residues inside the channel had to be removed through mutations to prevent repulsion with the negatively charged DNA. The second problem was addressed by using a special DNA polymerase to hold the DNA on top of the hemolysin and create a bottleneck.
The culmination of all these efforts was a publication two months back in which they could distinguish and sequence DNA strands ranging from 42 to 53 bases in length. That’s a good start, but of course it’s a trivial number compared to most respectable genomes of interest. And this is where the skeptics start chiming in. The error rate of the sequencing is currently 4%, mostly engendered by stalled DNA polymerase and bases that slip past without being read. For the three billion base pairs of the human genome, this will translate to about 120 million bases read incorrectly. That’s a staggering number, especially if you consider that many genetic disorders arise from changes in a single base pair. When you are dealing with complex disorders in which small changes in sequence mean the difference between health and disease, this kind of accuracy may just not be good enough. And of course, that’s assuming that such minor changes in sequence are truly meaningful, but that’s a different story. In addition there are probably going to be problems associated with the durability of the pore, not to mention the usual challenges involved in mass producing such a technology. I am assuming the midnight oil at Oxford is being burnt as we speak.
So nanopore sequencing may well be the next big thing. But its actual impact can only be judged after they iron out these key wrinkles and publish a few papers dealing with typical-sized genomes. Until then, cautious optimism without strenuous limb-flailing would be the correct response.
Image source
Read more at The Curious Wavefunction
Source: