| Visitors Now: | |
| Total Visits: | |
| Total Stories: |

| Story Views | |
| Now: | |
| Last Hour: | |
| Last 24 Hours: | |
| Total: | |
Dwave Adiabatic Quantum Computer used by Harvard to solve Protein folding problems
From
A team of Harvard University researchers, led by Professor Alan Aspuru-Guzik, have used Dwave's adiabatic quantum computer to solve a protein folding problem. The researchers ran instances of a lattice protein folding model, known as the Miyazawa-Jernigan model, on a D-Wave OneTM quantum computer.
We present the first quantum-mechanical implementation of lattice protein models using a programmable quantum device. We were able to encode and to solve the global minima solution for a small tetrapeptide and hexapeptide chain under several experimental schemes involving 5 and 8 qubits for the four-amino-acid sequence (Hydrophobic-Polar model) and 5, 27, 28, and 81 qubits experiments for the six-amino-acid sequence under the Miyazawa-Jernigan model for general pairwise interactions. For the experiment with 8 qubits, we simulated the dynamics of the quantum device with a Redfield equation with no adjustable parameters, obtaining excellent agreement with experiment. Since the quantum annealing algorithm not only finds the ground state but also the low-lying excited states, it provides information about the relevant minimum energy compact structures of protein sequences and it is useful to evaluate designability and stability such as that found in natural protein sequences, where the global minimum of free energy is well separated in energy from other misfolded states. The approach employed here can be extended to treat other problems in biophysics and statistical mechanics such as molecular recognition, protein design, and sequence alignment.
This was 81 qubit work. Dwave announced a 512 qubit chip was created late in 2011 and was being prepared for commercialization in 2012. The published research work generally lags the current work by 1-2 years. 512 qubits used for protein folding solutions would be a lot faster and should be able to solve far larger problems. Dwave should also be developing 2048 qubit chips in the next few years.
“The D-Wave computer found the ground-state conformation of six-amino acid lattice protein models. This is the first time a quantum device has been used to tackle optimization problems related to the natural sciences,” said Professor Alán Aspuru-Guzik from the Department of Chemistry and Chemical Biology at Harvard University.
Proteins contribute to virtually every process that occurs within a cell. The shape of a protein is closely related to its function. Understanding the shape of a protein helps researchers understand how it behaves, accelerating advances in many different areas of life sciences, including drug and vaccine design.
A cornerstone of computational biophysics, lattice protein folding models provide useful insight into the energy landscapes of real proteins. Understanding these landscapes, and how real proteins fold into the shapes that help give them their function, is an extremely difficult problem for today's computers to solve.
Dr. Alejandro Perdomo-Ortiz, the lead author of the paper, stated that: “Knowing that we can use real quantum computers to solve hard problems in biology is an exciting and important result. The techniques developed in this report can also be used to tackle other biophysical problems such as molecular recognition, protein design, and sequence alignment.”
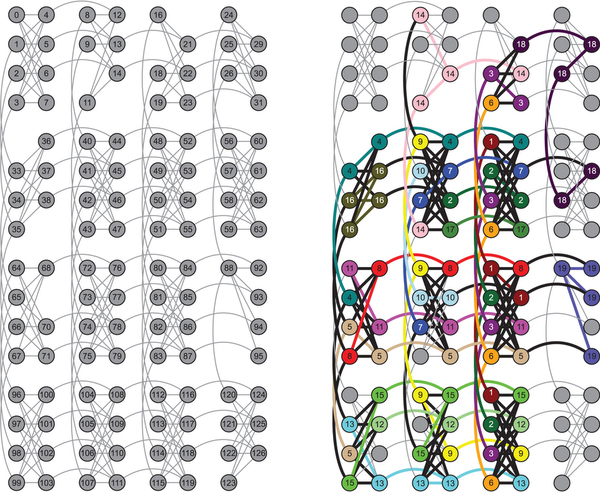
Device architecture and qubit connectivity. The array of superconducting quantum bits is arranged in 4 × 4 unit cells that consist of 8 quantum bits each. Within a unit cell, each of the 4 qubits in the left-hand partition (LHP) connects to all 4 qubits in the right-hand partition (RHP), and vice versa. A qubit in the LHP (RHP) also connects to the corresponding qubit in the LHP (RHP) of the units cells above and below (to the left and right of) it. (a) Qubits are labeled from 0 to 127 and edges between qubits represent couplers with programmable coupling strengths. Grey qubits indicate the 115 usable qubits, while vacancies indicate qubits under calibration which were not used. The larger experiments (Experiments 1,2, and 4) were performed on this chip, while the three remaining smaller experiments were run on other chips with the same architecture. (b) Embedding and qubit connectivity for Experiment 4, coloring the 81 qubits used in the experiment. Nodes with the same color represent the same logical qubit from the original 19-qubit Ising-like Hamiltonian resulting from the energy function associated with Experiment 4 (see Supplementary material for details). This embedding aims to fulfill the arbitrary connectivity of the Ising expression and allows for the coupling of qubits that are not directly coupled in hardware.
See more and subscribe to NextBigFuture at 2012-08-16 11:49:43 Source: http://nextbigfuture.com/2012/08/dwave-adiabatic-quantum-computer-used.html
Source: