| Online: | |
| Visits: | |
| Stories: |

| Story Views | |
| Now: | |
| Last Hour: | |
| Last 24 Hours: | |
| Total: | |
Natural GMOs Part 255. Complex modular architecture around a simple toolkit of wing pattern genes : Nature Ecology & Evolution
Modular evolution in the Heliconius butterfly follows mix and match gene shuffling rules:
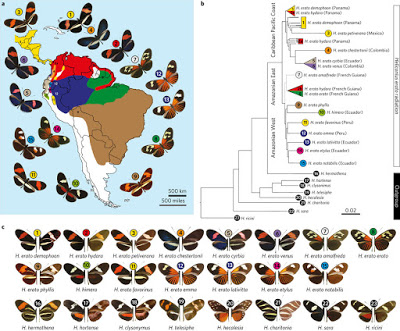
In Heliconius, conspicuous wing patterns are important for signalling toxicity to potential predators and play a role in mate selection . Natural selection favors Mìllerian mimicry among toxic butterflies, resulting in convergence between co-occurring species, as well as geographic divergence between populations of the same species 8 . Among Heliconius butterflies, the genetic basis of this wing diversity has been studied for nearly 60 years and more than 30 Mendelian loci have been described . Over the past decade, however, genetic research has shown that most of the complexity of colour variation across Heliconius is actually controlled by relatively few genes acting broadly across the fore- and hindwing…
Less than 0.2% of the genome was associated with wing pattern diversity across the H. erato radiation. This variation was highly modular and fell in non-coding regions near colour patterning genes, including optix, wntA and cortex, as well as a less well-documented colour pattern locus (Ro) that controls spatial variation of melanin in the upper forewing. Based on the proximity of these mostly non-coding intervals to known patterning genes, it is likely they represent cis-regulatory regions modulating the spatial expression of key patterning genes in discrete areas of the developing wing. In Heliconius, this modularity of cis-regulatory architecture provides a mechanism for rapid evolution of novel morphologies.
Both shuffling of existing modules and de novo evolution of new modules is associated with phenotypic diversity in H. erato. Indeed, we can recreate the colour pattern diversity across the H. erato radiation using a combination of non-genic regions near four colour pattern genes (Fig. 6). This conclusion is perhaps best exemplified in the distribution of genetic variation around wntA, where different colour pattern races have different combinations of four distinct genomic intervals. These different intervals are likely to regulate the expression of wntA in different areas of the forewing to adjust the position, size and shape of the forewing band to closely match patterns in other co-occurring warningly coloured butterfly species. Within this modular framework, recombination can reshuffle existing regulatory variation to generate new combinations of regulatory elements and new wing pattern phenotypes. Recombination of colour pattern modules and introgression into other populations is likely to be driven by high rates of gene flow between adjacent populations. For example, H. e. amalfreda appears to have evolved via recombination of regulatory variation between rayed (H. e. erato) and red-banded (H. e. hydara) haplotypes that instantaneously generated a novel wing pattern, a process that closely mirrors the one recently described in the co-mimetic forms of H. melpomene .
New regulatory modules associated with wing pattern variation can also evolve de novo, further increasing the flexibility of these regions to generate pattern diversity. This was evident in the independent evolution of the yellow hindwing bar in the H. erato clade (Fig. 5), and also in the comparison of regulatory variation around the red patterning locus between H. erato and its co-mimic H. melpomene. Red pattern variation in the two species is similarly generated by regulatory differences at the optixlocus , and the genomic position and order of its cis-regulatory elements is broadly similar . Furthermore, in both species distinct intervals were associated with different red pattern elements, and ‘enhancer shuffling’ through recombination has similarly generated novel red pattern phenotypes . This implies considerable conservation of function of optix cis-regulatory regions that were re-used to generate the convergent patterns that underlie mimicry…
Full text @Complex modular architecture around a simple toolkit of wing pattern genes : Nature Ecology & Evolution:
Source: http://gmopundit.blogspot.com/2017/01/natural-gmos-part-255-complex-modular.html